CRISPR Resources
General resources
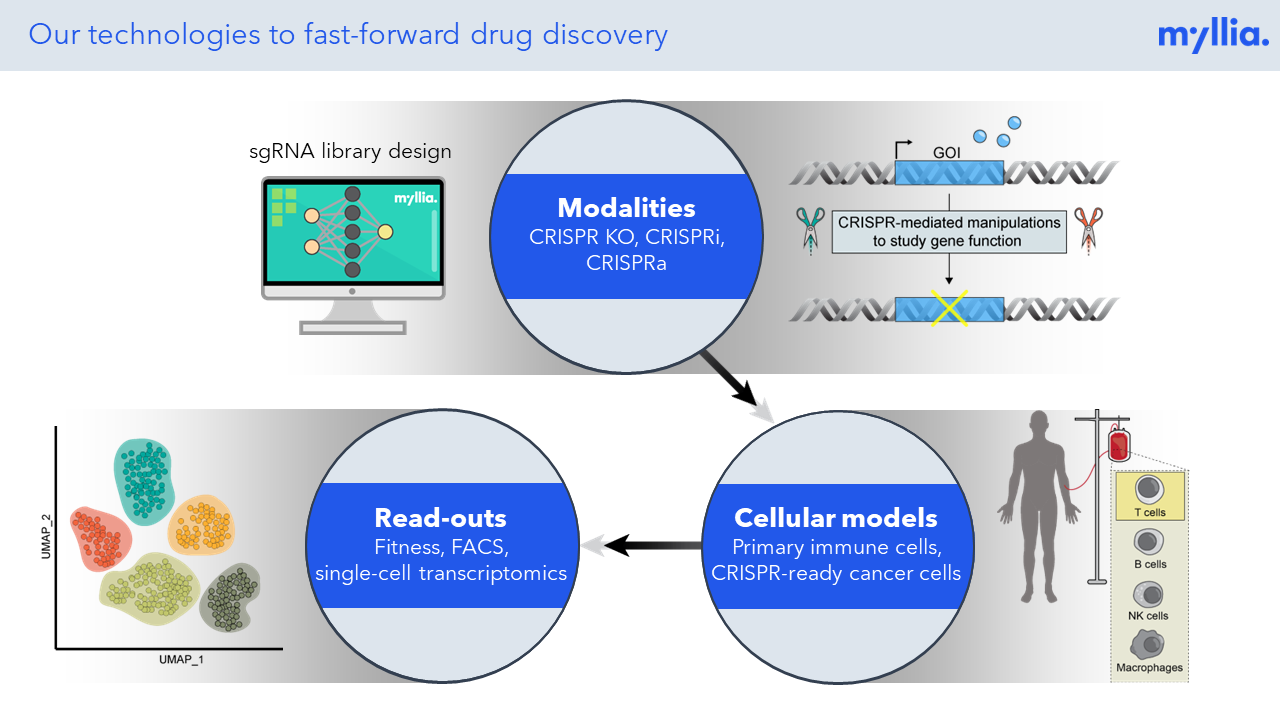
Our Factsheet
Check out Myllia's technology platform enabling drug target discovery and functional genomics at single-cell resolution
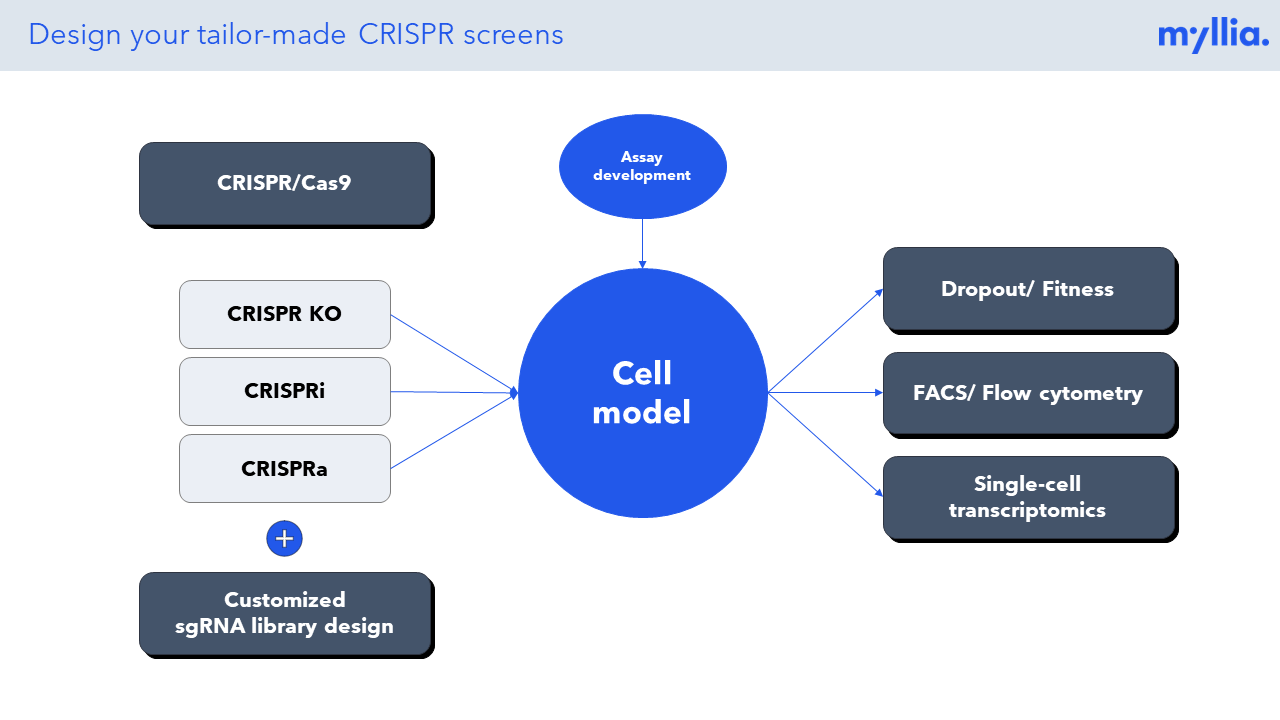
Our Portfolio
At Myllia, pooled CRISPR screens are performed at large scale, and our unique platform involves a broad portfolio of CRISPR/Cas9 technologies

CRISPR-ready cancer cell lines for CRISPRko & CRISPRi screens
Browse through our catalogue of CRISPR-ready cancer cell lines for your next tailor-made CRISPR screen

Next-generation CRISPR screening for drug target discovery
De-risk target discovery and drug development using the best possible validated targets

CRISPR screens for immuno-oncology (IO) research
Utilize our tailor-made CRISPR screens to characterize your immuno-oncology drug candidates and CAR-T cell products
Posters
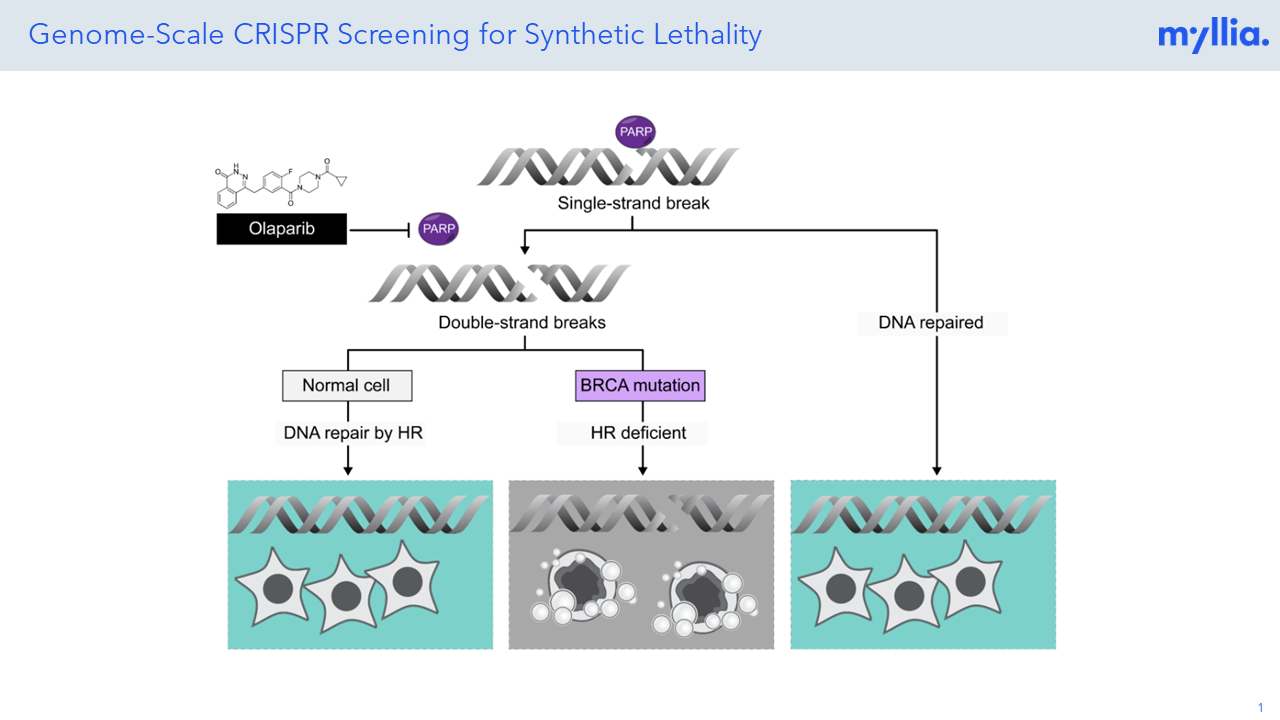
Poster: A Genome-Scale PARPi Screen for Synthetic Lethality
At Myllia, we’ve performed a genome-scale CRISPR KO screen in HeLa cells to identify novel or well-known genes that are involved in DNA damage repair and would resemble gene pairs with PARP.
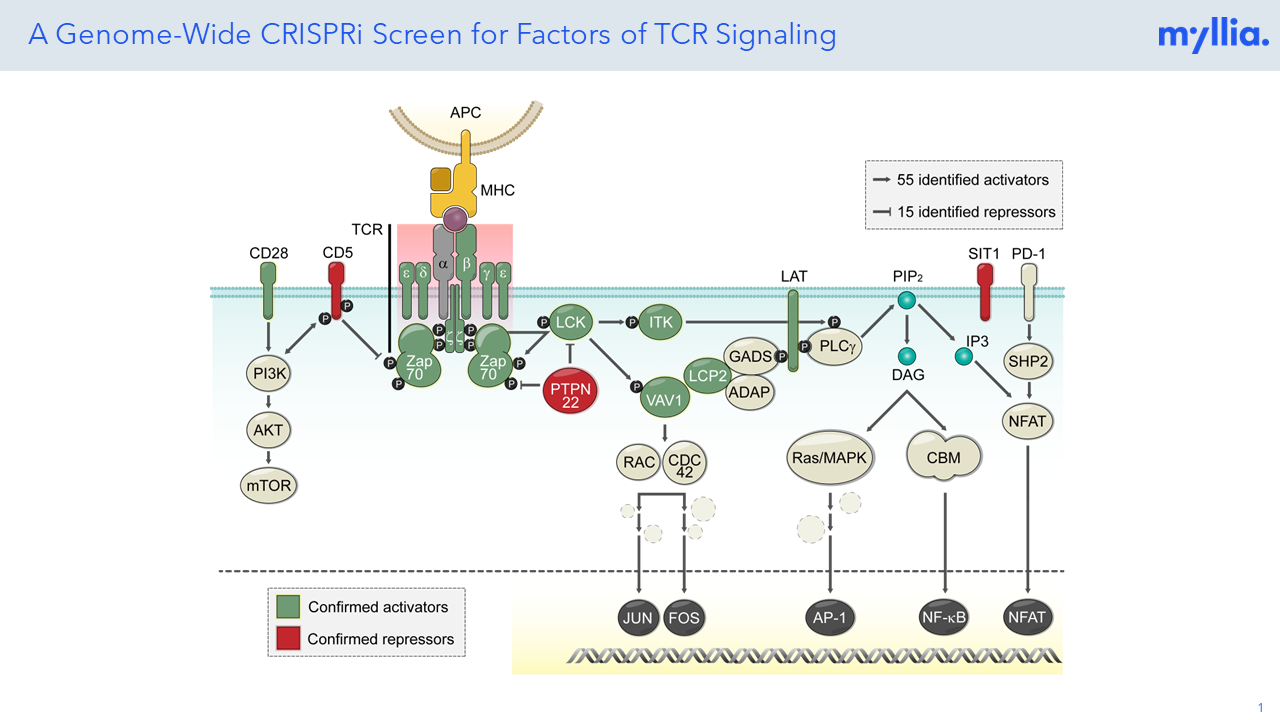
Poster: A Genome-Wide CROP-Seq Screen Reveals Mediators of T Cell Signaling
The first-of-its-kind genome-scale CRISPRi screen was conducted to verify factors involved in TCR signaling pathways. Unravelling these T cell-intrinsic molecular factors could potentially unlock new therapeutic targets and strategies for modulating T cell responses in various immune-related disorders.
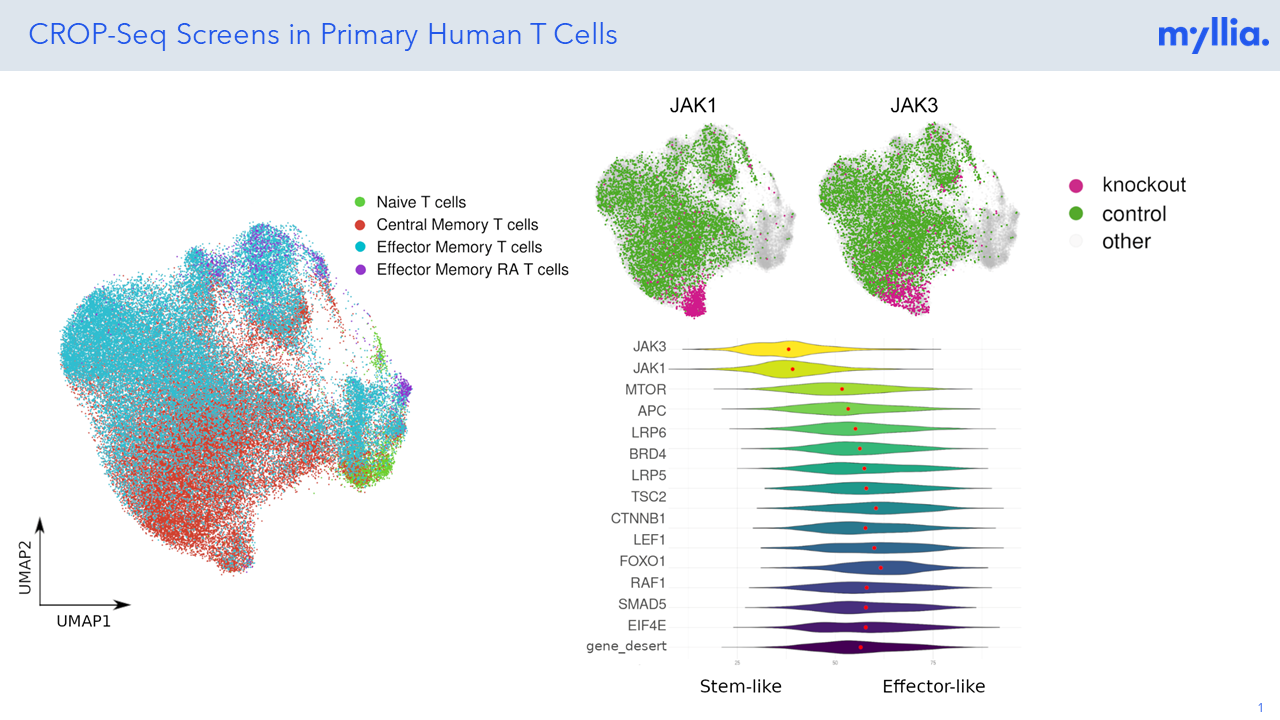
Poster: Large-Scale Single-Cell CRISPR Screens in Primary Human T Cells
The CROP-seq experiment in pan T-cells involved a targeted single-cell RNA sequencing of 300 mRNAs. At Myllia, we can identify rare subsets, transitional states, and distinct functional profiles that may be masked in bulk analyses, providing a more comprehensive understanding of T cell activation and differentiation.
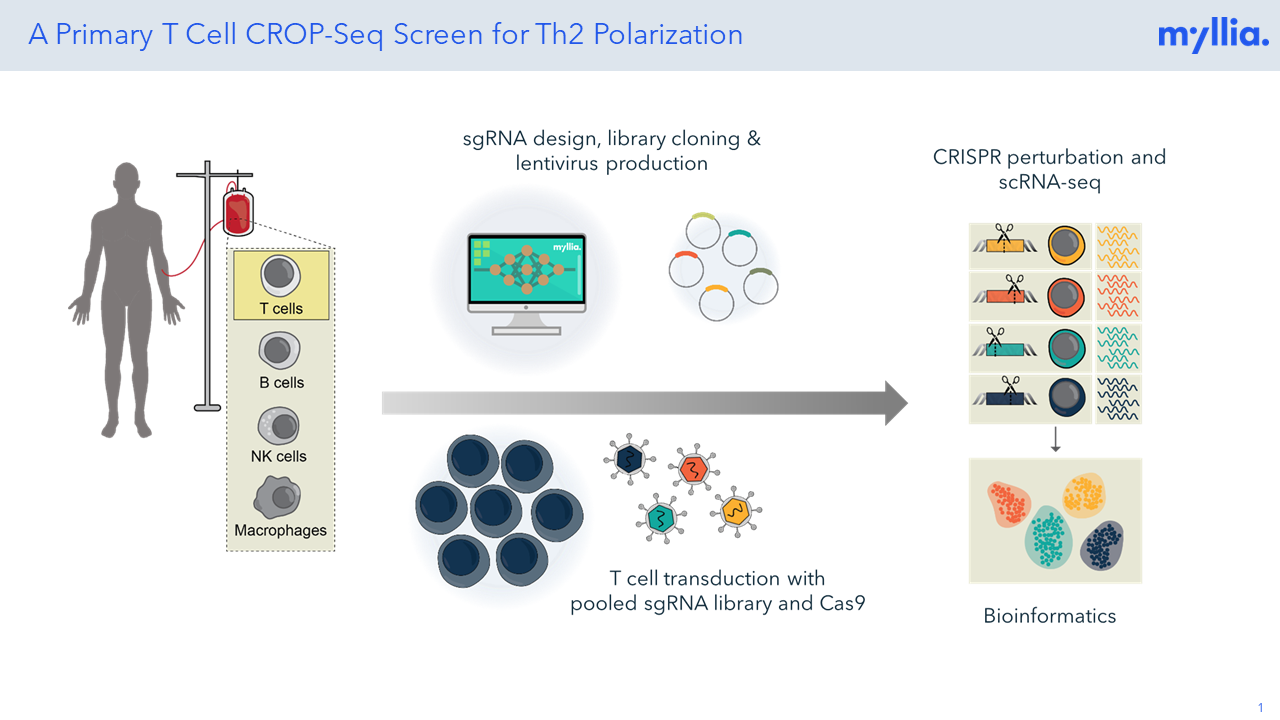
Poster: Single-Cell CRISPR Screens in Primary Human T Cells Identify Regulators of Th2 Cell Skewing
T helper 2 (Th2) cell differentiation plays a critical role in regulating allergic responses, defense against parasites, and immune homeostasis. In a CROP-Seq experiment with CD4+ T cells, Th2-skewed cells were studied using a sgRNA library targeting 102 genes with a targeted read-out of 300 mRNAs.
Slide decks

A Genome-Scale PARPi CRISPR KO Screen for Synthetic Lethality
CRISPR KO screen at genome-scale in HeLa cells to identify novel or well-known genes that are involved in DNA damage repair to explore PARPi synthetic lethality gene pairs

A Genome-Wide CROP-Seq CRISPRi Screen for Factors of TCR Signaling
At Myllia, the Perturb-seq (CROP-seq) workflow has been adapted to enable genome-scale CRISPRi (CRISPR interference) screens in Jurkat cells at single-cell resolution. The first-of-its-kind genome-scale CRISPRi screen was conducted to verify factors involved in TCR signaling pathways.

Large-Scale CROP-Seq Screening in Primary Human T Cells
At Myllia, we can identify rare subsets, transitional states, and distinct functional profiles that may be masked in bulk analyses, providing a more comprehensive understanding of T cell activation and differentiation. The CROP-seq experiment in pan T-cells involved a targeted single-cell RNA sequencing of 300 mRNAs.

A CROP-Seq Screen for Modulators of Th2 Cell Polarization
In a CROP-Seq experiment with CD4+ T cells, Th2-skewed cells were studied using a sgRNA library targeting 102 genes with a targeted read-out of 300 mRNAs.

Pooled CRISPR screens for drug target discovery
At Myllia, we use the CROP-Seq technology to perturb cells with CRISPR and profile transcriptional outcomes by RNA sequencing at single-cell resolution.
Videos

About us
Discover the unique CRISPR screening portfolio developed at Myllia Biotechnology

How do CRISPR screens work?
Explore how we can use CRISPR screens to support drug target identification

CRISPR/Cas9-based screens: CRISPRko vs. CRISPRi
Learn more about the differences of Cas9-based CRISPRko (knockout) and CRISPRi (interference) screens

CROP-Seq: Single-cell CRISPR screening
Learn how the CROP-Seq technology at Myllia can help to elucidate novel drug targets at single-cell resolution

Targeted sequencing
Would you like to know more about targeted sequencing applications? Explore how tailored PCR panels can boost the NGS read-outs of your next CRISPR screens

Primary human T cell CRISPR screening
At Myllia, CRISPR screens are performed in primary human T cells to explore factors of T cell activation, differentiation and exhaustion. Learn more about our CROP-seq technology to boost CAR-T and TCR-T cell potency!
