Large-Scale CROP-Seq Screening in Primary Human T Cells
At Myllia, we can identify rare subsets, transitional states, and distinct functional profiles that may be masked in bulk analyses, providing a more comprehensive understanding of T cell activation and differentiation. The CROP-seq experiment in pan T-cells involved a targeted single-cell RNA sequencing of 300 mRNAs.
T cell plasticity refers to the ability of T cells to adopt different phenotypes in response to various signals, allowing them to exhibit diverse immune functions. However, the factors that determine the stability and reversibility of these phenotypic transitions are not fully understood.
The Screen
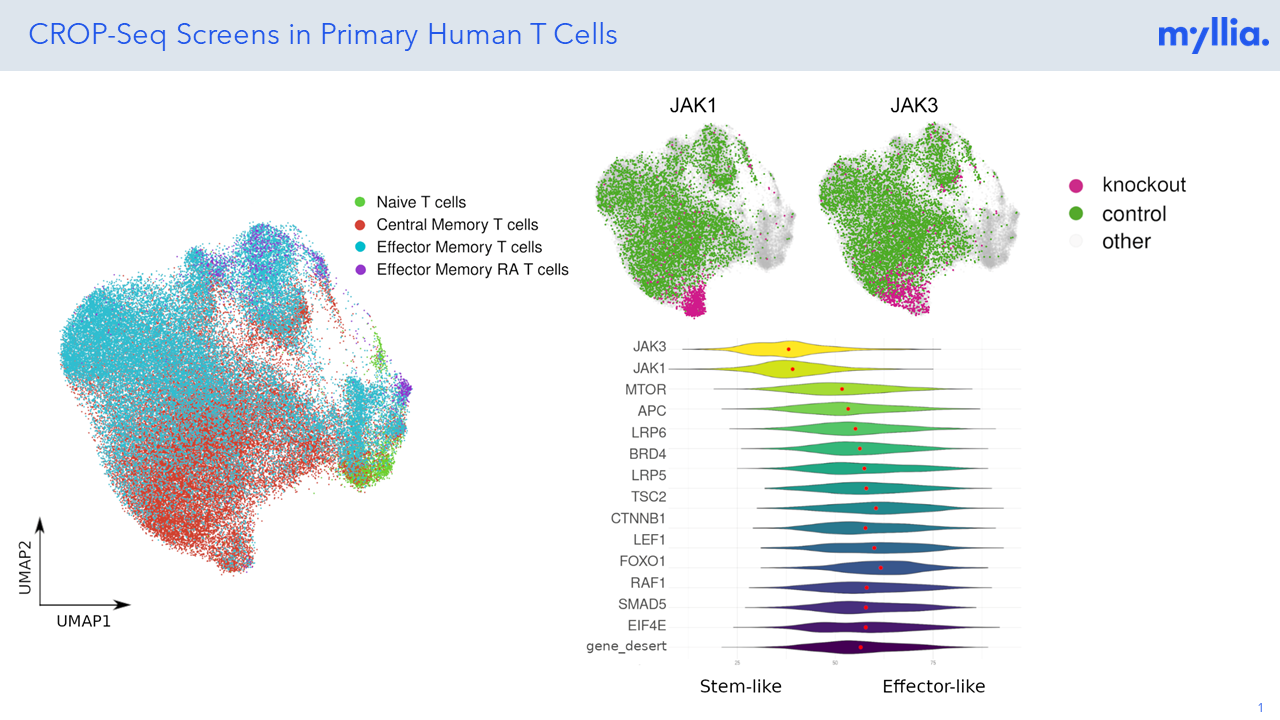
The single-cell resolution allows the examination of individual T cells within a heterogeneous population, unveiling cellular diversity and functional heterogeneity. At Myllia, we can identify rare subsets, transitional states, and distinct functional profiles that may be masked in bulk analyses, providing a more comprehensive understanding of T cell activation and differentiation. The CROP-seq experiment in pan T-cells involved a targeted single-cell RNA sequencing of 300 mRNAs, which included T cell activation markers, markers for different cell types, cell cycle markers, and immune checkpoint genes.
Results & Conclusions
Our screens revealed that knockouts of JAK1 and JAK3 resulted in a significant increase in the expression of TNFRSF9, also known as 4-1BB, which is a checkpoint gene associated with immune regulation and T cell activation, suggesting an important role of JAK-STAT signaling in both CD4+ and CD8+ T cell subsets. Leveraging our know-how on T cell subsets and transcriptomic markers, we can assess T cell plasticity at large scale and providing insights into functions of genes linked to immune pathways.
Submit your credentials to download the full slide deck and explore our primary human T cell CRISPR screening platform!
