OUR TECHNOLOGY PLATFORM ENABLING CRISPR SCREENS AT SINGLE-CELL RESOLUTION
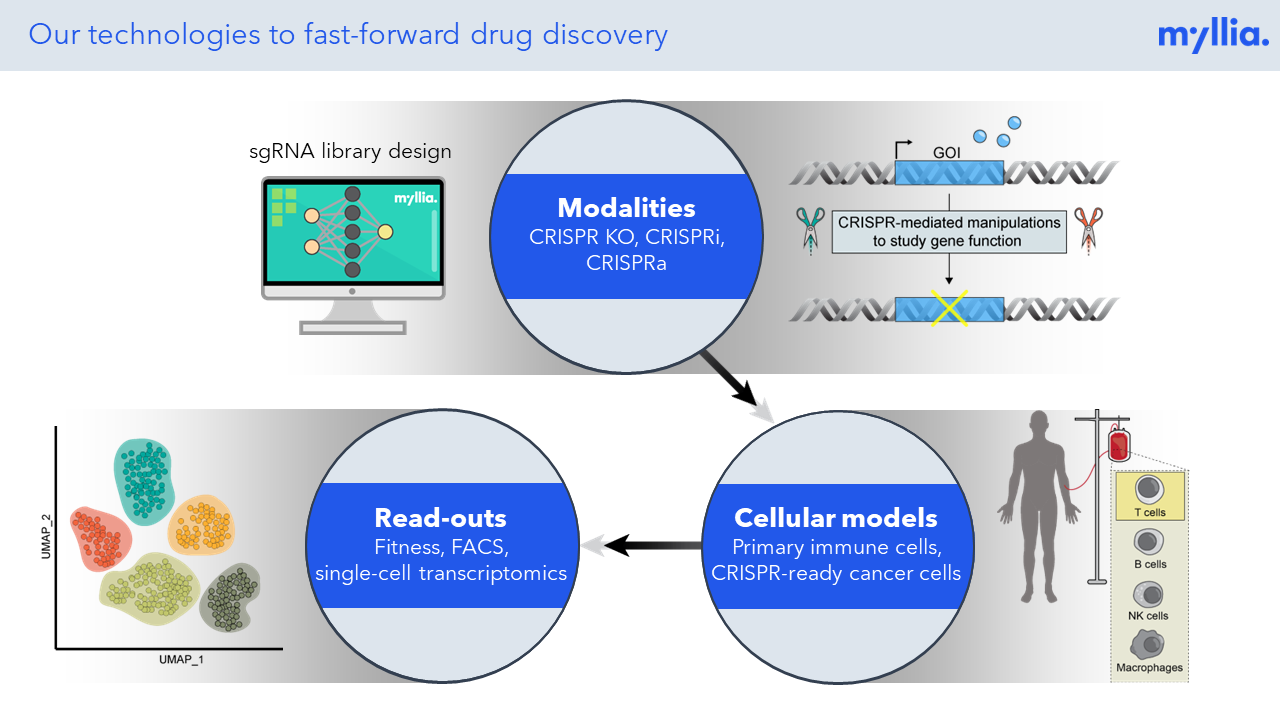
Myllia Biotechnology combines CRISPR screening with single-cell RNA sequencing, leveraging two transformative technologies to enable genetic screening for complex phenotypes.
CRISPR Screening
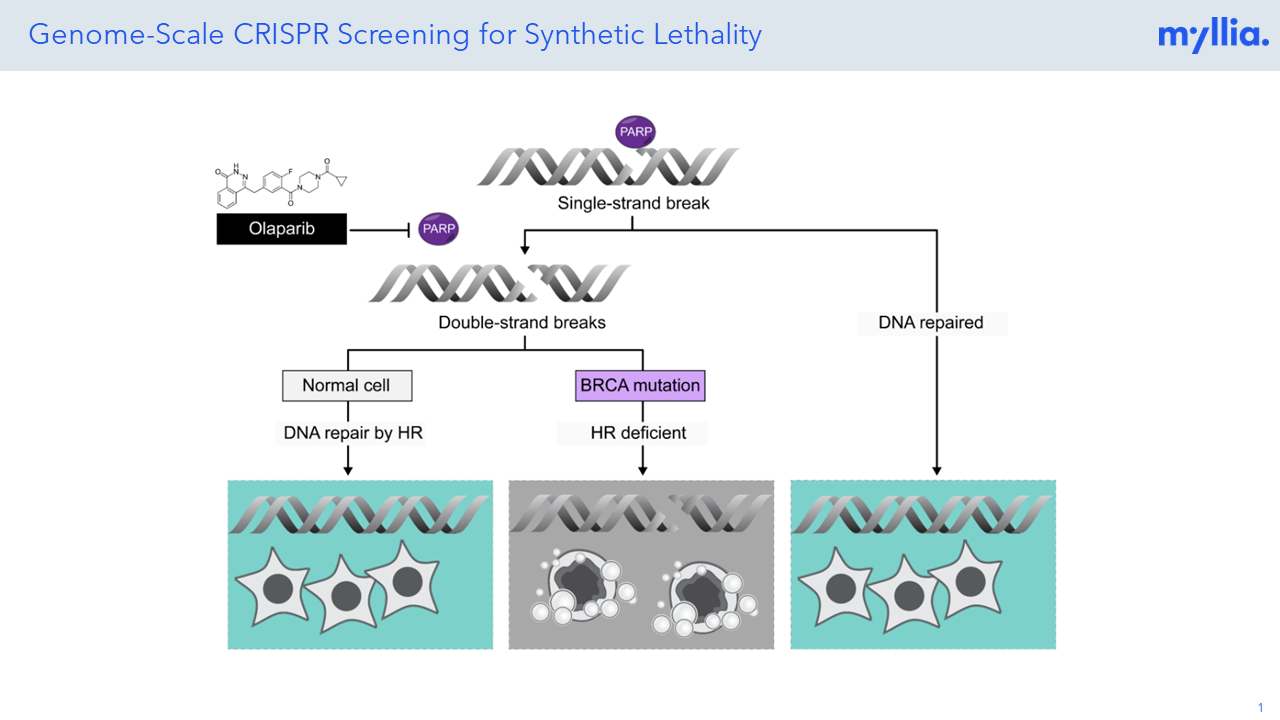
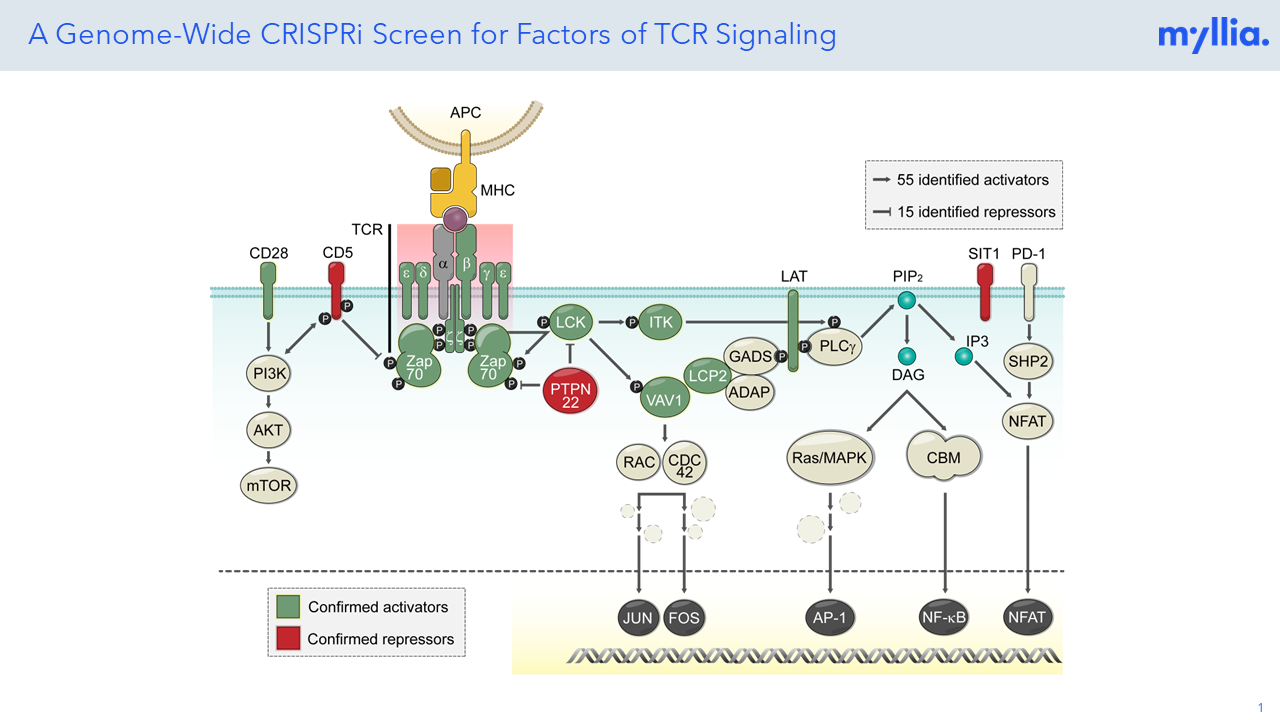
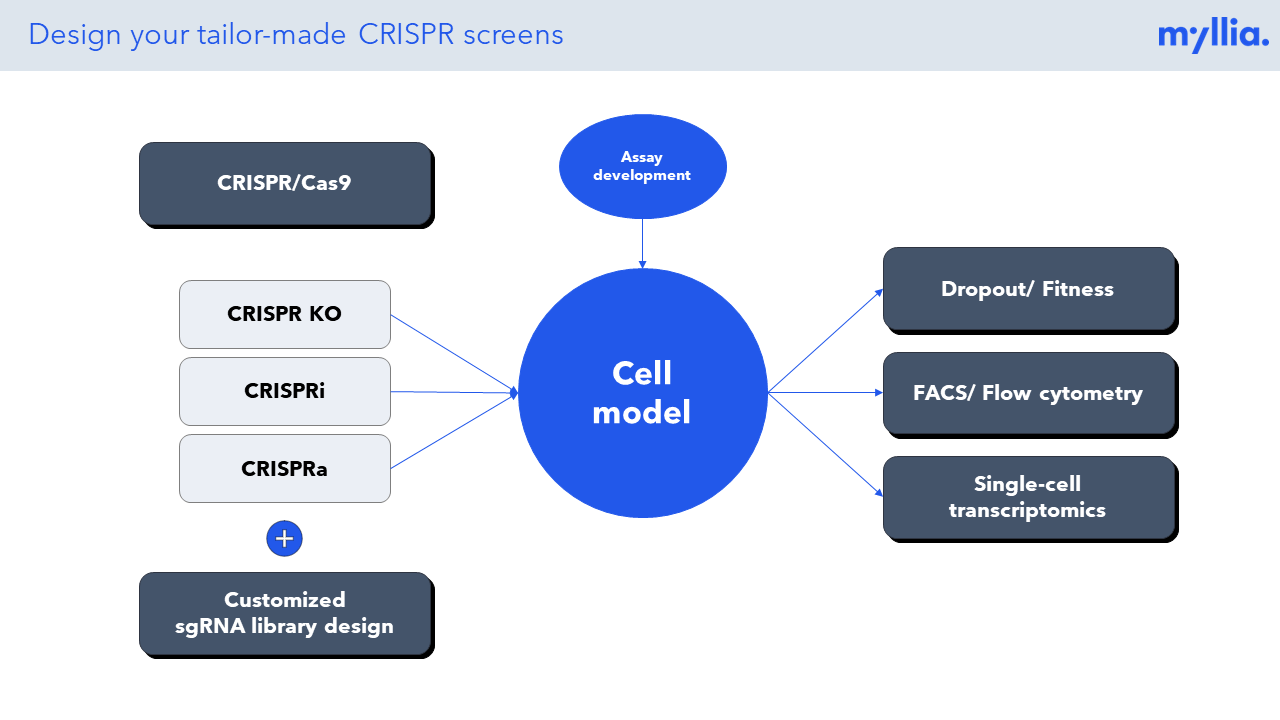
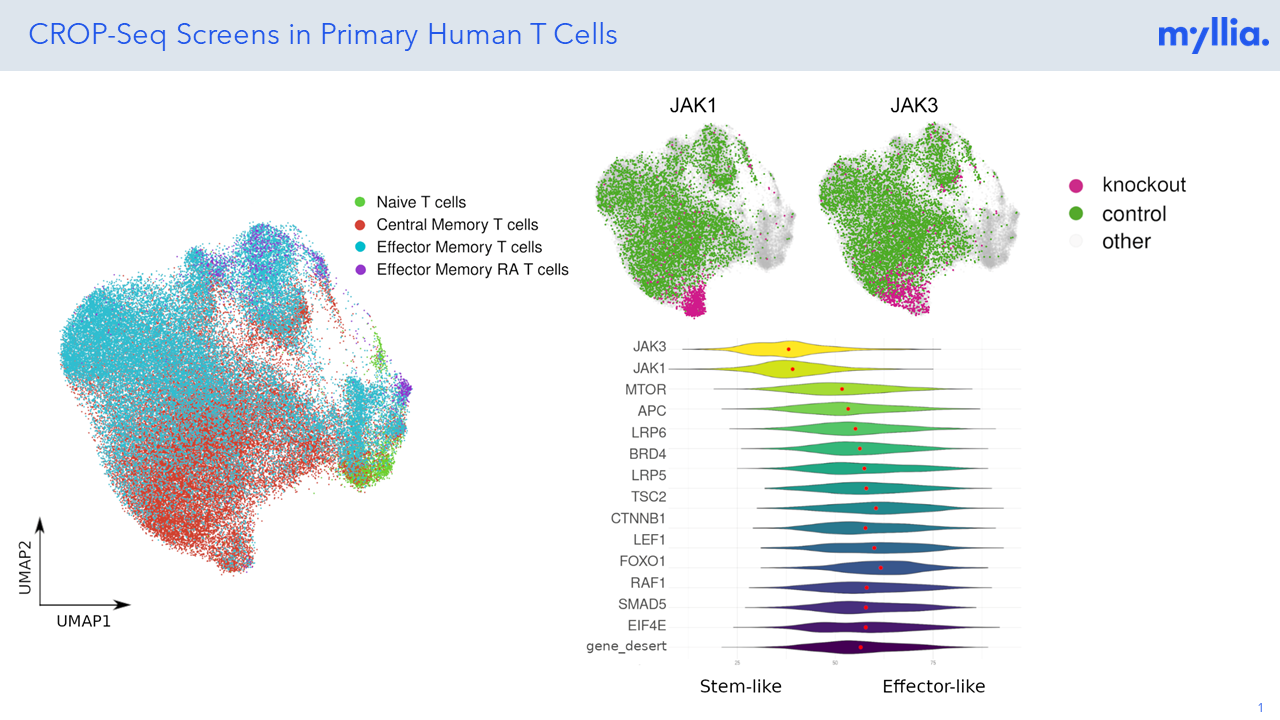
CRISPR screening has revolutionized the unbiased annotation of gene function, but most screens done so far have been confined to rather simplistic read-outs (usually life/death of the target cells). Combining CRISPR perturbation with single-cell sequencing allows researchers to assess much more complex phenotypes, thus effectively broadening the scope of these screens. At Myllia, we use the CROP-Seq technology to perturb cells with CRISPR and profile transcriptional outcomes by RNA sequencing at single-cell resolution. Importantly, our technology is applicable across a wide range of cell types including primary cells. The latter include primary human T cells which are of great interest for the discovery of novel targets in immuno-oncology.
CRISPR Screening
CRISPR screening has revolutionized the unbiased annotation of gene function, but most screens done so far have been confined to rather simplistic read-outs (usually life/death of the target cells). Combining CRISPR perturbation with single-cell sequencing allows researchers to assess much more complex phenotypes, thus effectively broadening the scope of these screens. At Myllia, we use the CROP-Seq technology to perturb cells with CRISPR and profile transcriptional outcomes by RNA sequencing at single-cell resolution. Importantly, our technology is applicable across a wide range of cell types including primary cells. The latter include primary human T cells which are of great interest for the discovery of novel targets in immuno-oncology.
CAS9-EXPRESSING CELL LINE
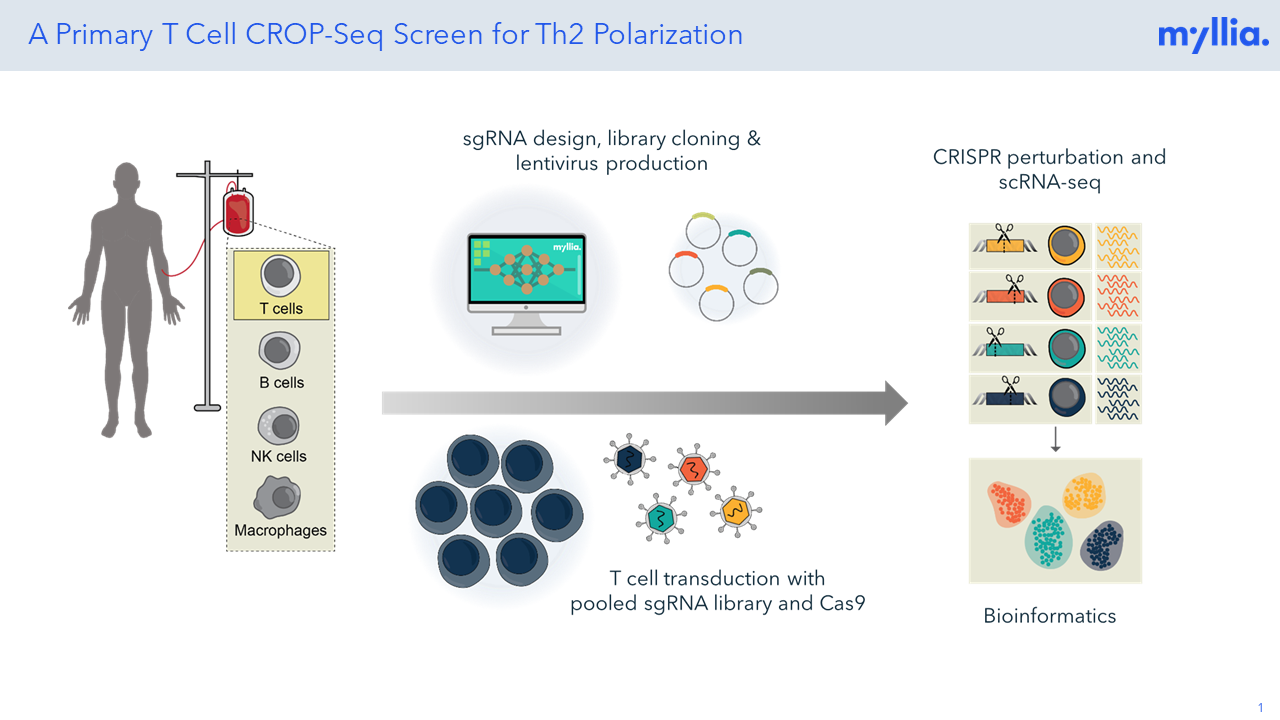
Every CRISPR screen starts in a cell line that harbors Cas9.



LENTIVIRAL GUIDE RNA LIBRARY
Cells are infected with a pooled lentiviral single-guide RNA (sgRNA) library.



CRISPR PERTURBATIONS
Following perturbation with Cas9 and a suitable guide RNA, every single cell in the pool will carry a knockout for a different gene.



Single-cell RNA sequencing
The CROP-Seq (“CRISPR droplet sequencing”) technology measures transcriptome responses to CRISPR perturbation. It offers the flexibility of arrayed CRISPR screens at the scale of pooled CRISPR screens, thus providing a synergy of the two widely popular screening paradigms.
Single-cell RNA sequencing
The CROP-Seq (“CRISPR droplet sequencing”) technology measures transcriptome responses to CRISPR perturbation. It offers the flexibility of arrayed CRISPR screens at the scale of pooled CRISPR screens, thus providing a synergy of the two widely popular screening paradigms.
SINGLE CELL LIBRARY PREPARATION
Each single cell is then encapsulated in a lipid droplet together with a barcoded bead. Reverse transcription occurs on the surface of the bead, thus creating a unique transcriptomic fingerprint for each cell.



GUIDE RNA MAPPING
Mapping of the guide RNAs will connect each single-cell transcriptome to the guide RNA perturbation that caused the transcriptomic phenotype.



NGS AND BIOINFORMATIC ANALYSIS
Single-cell sequencing datasets are typically large and complex. We are routinely analyzing these and are providing analyses that are customized to the needs of our clients.