A Genome-Scale PARPi CRISPR KO Screen for Synthetic Lethality
CRISPR KO screen at genome-scale in HeLa cells to identify novel or well-known genes that are involved in DNA damage repair to explore PARPi synthetic lethality gene pairs
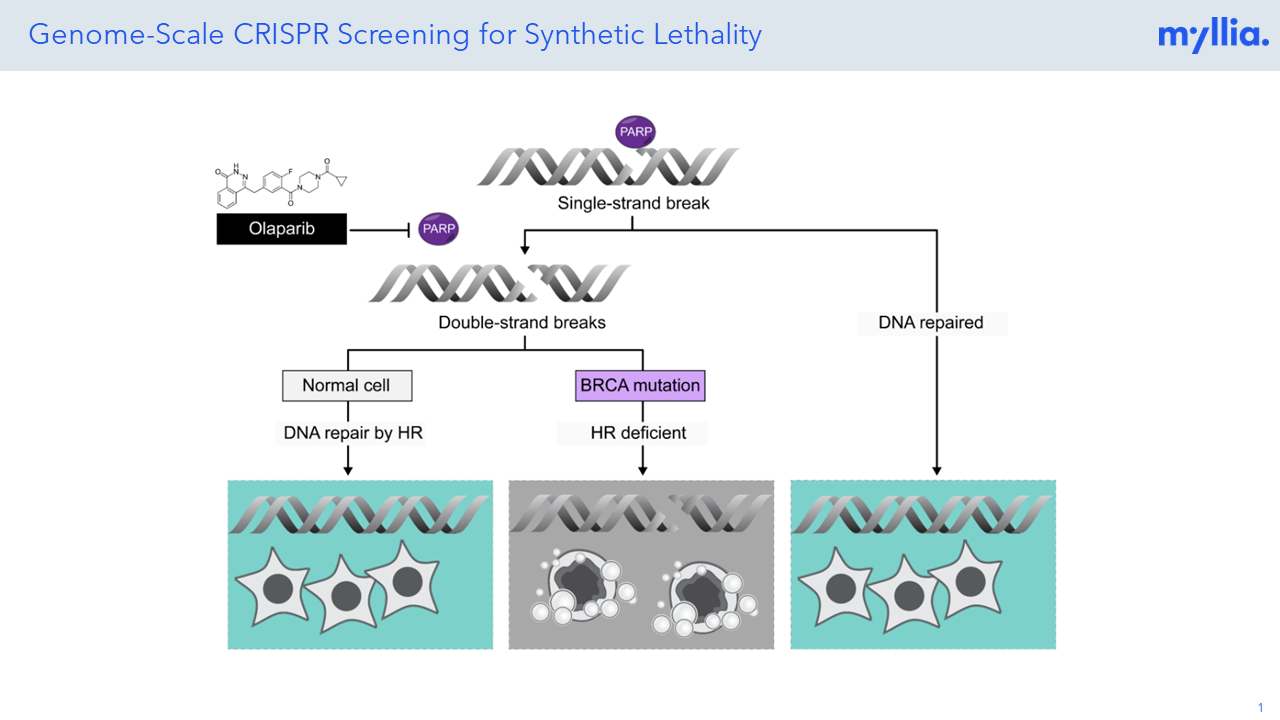
Synthetic lethality has emerged as a promising approach in cancer research, aiming to exploit specific genetic vulnerabilities in tumor cells. Despite its potential, the identification and validation of synthetic lethal gene pairs remain challenging due to the difficulty in accurately predicting and prioritizing candidate gene pairs for experimental testing, given the complexity of the cancer genome and the vast number of potential interactions.
The Screen
PARP (Poly-ADP ribose polymerase) inhibition is a therapeutic strategy used in cancer treatment aiming to prevent cancer cells from efficiently repairing their DNA, thus leading to genomic instability and cell death, particularly in tumors with BRCA1/2 mutations. At Myllia, we’ve performed a genome-scale CRISPR KO screen in HeLa cells to identify novel or well-known genes that are involved in DNA damage repair and would resemble gene pairs with PARP.
Results & Conclusion
Our results indicate that most identified genes are associated with different DDR mechanisms, including BER, ICL repair, homologous recombination (HR), and NHEJ or Alt-NHEJ pathways. Comparing our hit list with data derived from 23 publicly available screens, we can confirm a total of 109 hits across 14 different cell lines, hence demonstrating how synthetic lethality CRISPR screens can support target identification for novel combination therapies.
Submit your credentials and explore our capabilities and functional read-outs!
